Adding extra metadata via CSV/TSV¶
Auspice datasets can define a number of colorings – e.g. “country”, “age” – which you can switch between via the dropdown in the left-hand sidebar. By dragging CSV / TSV files onto the browser you can add in extra colorings. These extra data are processed within the browser, so no information leaves the client, which can be useful for viewing private metadata.
Format¶
The first column defines the names of the strains / samples in the tree, while the first row (header row) defines the metadata names.
The separator can be either a tab character or a comma & the file extension should be .tsv or .csv, respectively.
You can add as many columns you want, each will result in a different coloring of the data being made available
TSV Example:
strain secret
USVI/19/2016 A
USVI/28/2016 B
USVI/41/2016 C
USVI/42/2016 C
How to use¶
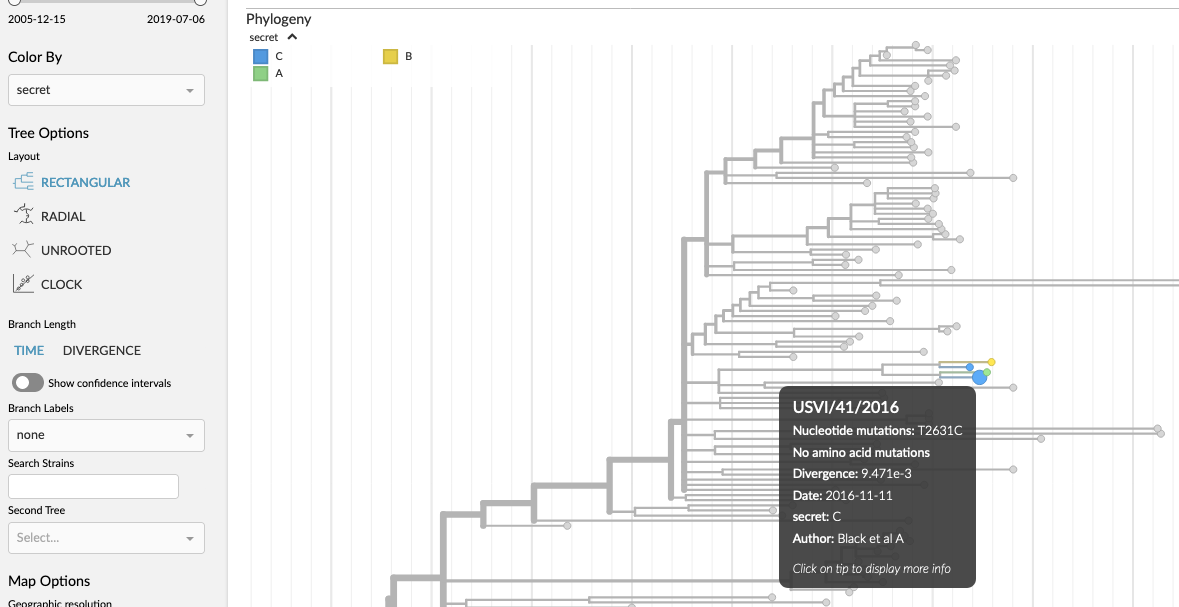
After creating the TSV/CSV file, you simply drag the file onto the browser (which is running nextstrain or auspice). You will see a notification pop up in the top-right, and (as long as things worked as they should), a new entry will appear in the “Color By” dropdown in the sidebar.
For instance, if we save the above example as a TSV file and drag it onto the browser running nextstrain.org/zika we will create a new color-by called “secret”. This allows you to change the coloring (via the drop-down in the sidebar) to “secret” to reveal the information attached to those 4 USVI strains. The tree will now look like:
Gotchas¶
The type of the data is currently always categorical – i.e. numeric data will work, but won’t be very usable if there are many values.
Any of following columns specified in the CSV/TSV will be ignored: “name”, “div”, “num_date”, “vaccine”, “labels”, “hidden”, “mutations”, “url”, “authors”, “accession”, “traits”, “children”.