Curate data from GISAID search and downloads
The following instructions describe how to curate data for a region-specific analysis (e.g., identifying recent introductions into Washington State) using GISAID’s “Search” page and curated regional data from the “Downloads” window. Inferences about a sample’s origin strongly depend on the composition of your dataset. For example, discrete trait analysis models cannot infer transmission from an origin that is not present in your data. We show how to overcome this issue by adding previously curated contextual sequences from Nextstrain to your region-specific dataset.
Table of Contents
Login to GISAID
Navigate to GISAID (gisaid.org) and select the “Login” link.

GISAID homepage with login link
Login to your GISAID account. If you do not have an account yet, register for one (it’s free) by selecting the “Registration” link.
GISAID login page with registration link
Select “EpiCoV” from the top navigation bar.

GISAID navigation bar with “EpiCoV” link
Search for region-specific data
Select “Search” from the EpiCoV navigation bar.

GISAID EpiCoV navigation bar with “Search” link
Find the “Location” field and start typing “North America /”. As you type, the field will suggest more specific geographic scales.
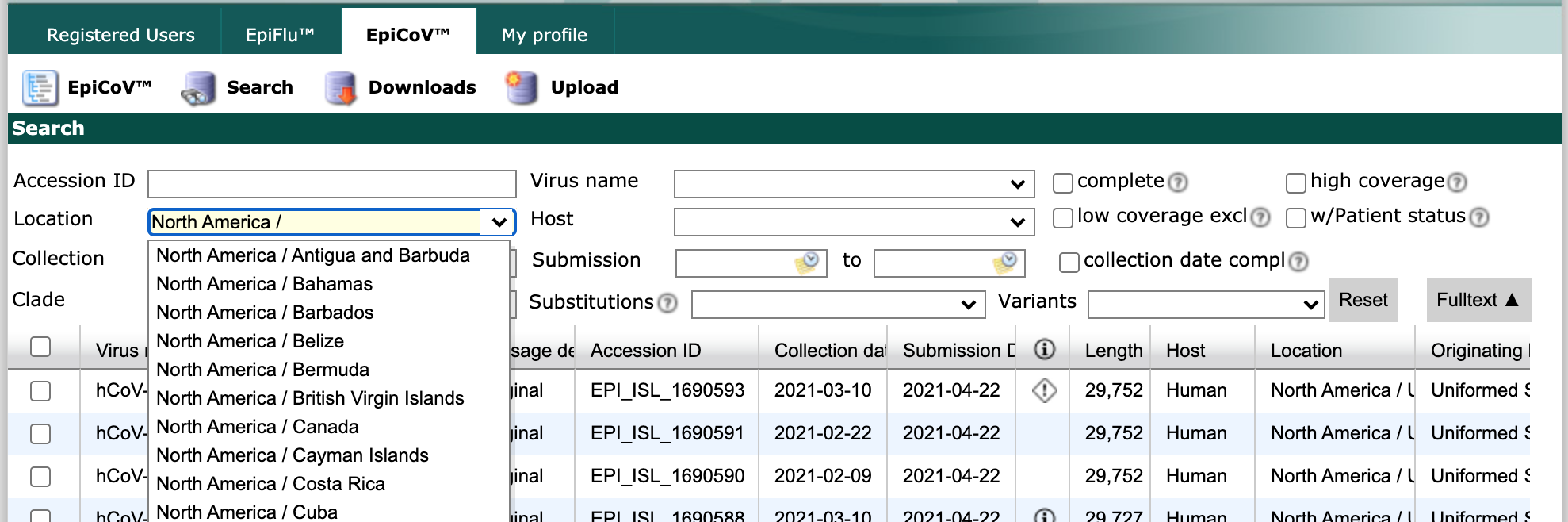
GISAID initial search interface
Finish by typing “North America / USA / Washington”. Select all strains collected between May 1 and June 1 with complete genome sequences and collection dates. Click the checkbox in the header row of the results display, to select all strains that match the search parameters.
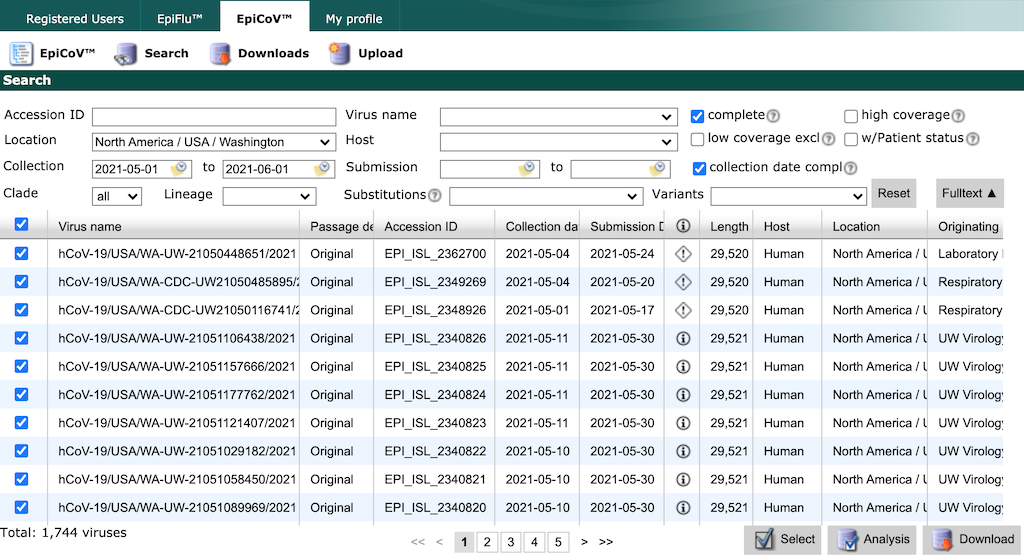
GISAID search results for “Washington”
Warning
GISAID limits the number of records you can download at once to 5000. If you need to download more records, constrain your search results to smaller windows of time by collection date and download data in these smaller batches.
Select the “Download” button in the bottom right of the search results. There are two options to download data from GISAID, both of which we describe below.
Option 1: Download “Input for the Augur pipeline”
From the resulting “Download” window, select “Input for the Augur pipeline” as the download format.
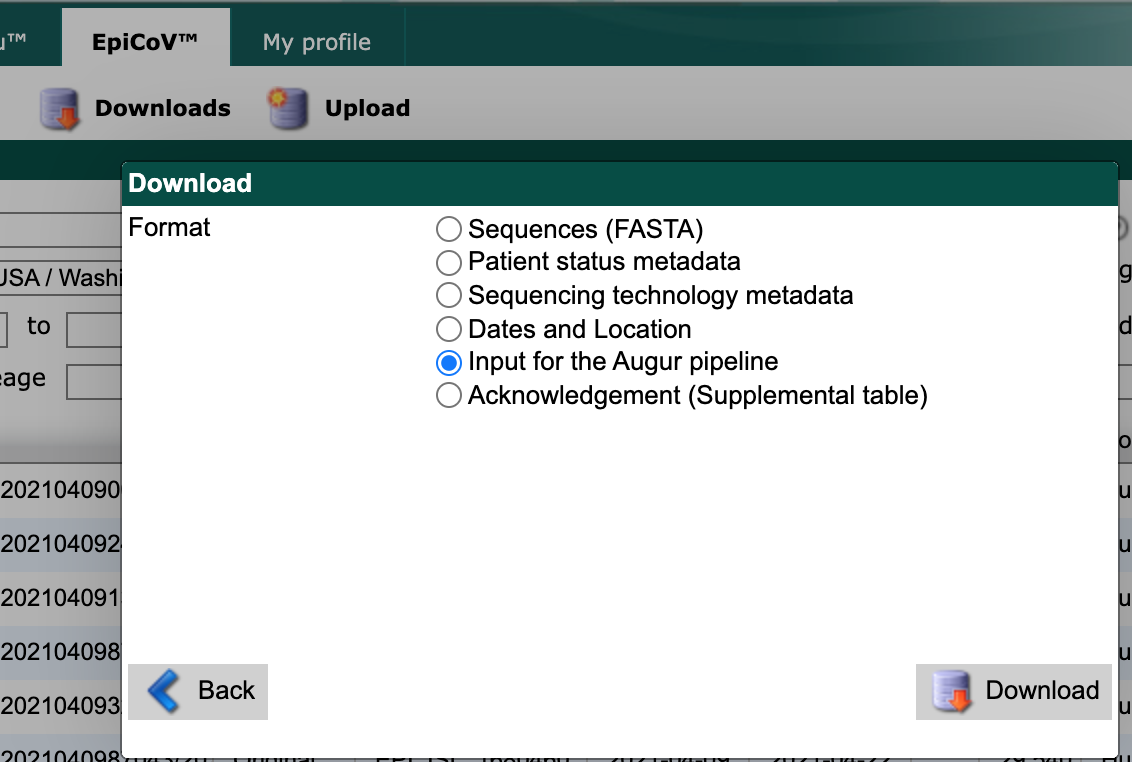
GISAID search download window showing “Input for the Augur pipeline” option
Select the “Download” button and save the resulting file to the data/ directory with a descriptive name like gisaid_washington.tar. This tar archive contains compressed metadata and sequences named like 1622567829294.metadata.tsv.xz and 1622567829294.sequences.fasta.xz, respectively.
You can use this tar file as an input for the Nextstrain workflow, as shown below. The workflow will extract the data for you. Create a new workflow config file, in the top-level of the ncov directory that defines your analysis or “builds”.
# Define inputs for the workflow.
inputs:
- name: washington
# The workflow will detect and extract the metadata and sequences
# from GISAID tar archives.
metadata: data/gisaid_washington.tar
sequences: data/gisaid_washington.tar
Next, you can move on to the heading below to get contextual data for your region of interest. Alternately, you can extract the tar file into the data/ directory prior to analysis.
tar xvf data/gisaid_washington.tar
Rename the extracted files to match the descriptive name of the original archive.
mv data/1622567829294.metadata.tsv.xz data/gisaid_washington_metadata.tsv.xz
mv data/1622567829294.sequences.fasta.xz data/gisaid_washington_sequences.fasta.xz
You can use these extracted files as inputs for the workflow.
# Define inputs for the workflow.
inputs:
- name: washington
# The workflow also accepts compressed metadata and sequences
# from GISAID.
metadata: data/gisaid_washington_metadata.tsv.xz
sequences: data/gisaid_washington_sequences.fasta.xz
Option 2: Download “Sequences” and “Patient status metadata”
Alternately, you can download sequences and metadata as two separate uncompressed files. First, select “Sequences (FASTA)” as the download format. Check the box for replacing spaces with underscores. Select the “Download” button and save the resulting file to the data/ directory with a descriptive name like gisaid_washington_sequences.fasta.
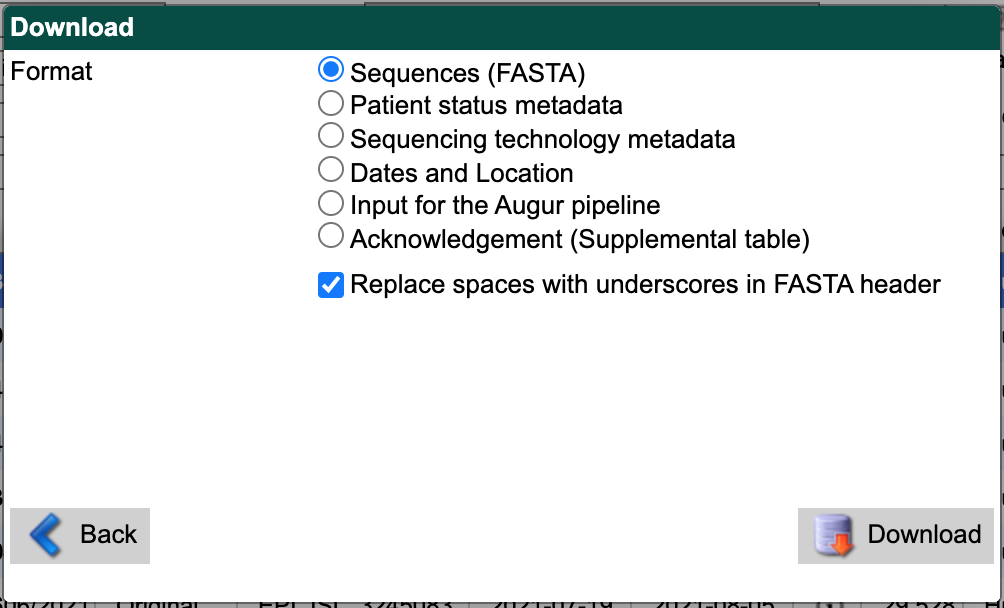
GISAID search download window showing “Sequences (FASTA)” option
From the search results interface, select the “Download” button in the bottom right again. Select “Patient status metadata” as the download format. Select the “Download” button and save the file to data/ with a descriptive name like gisaid_washington_metadata.tsv.
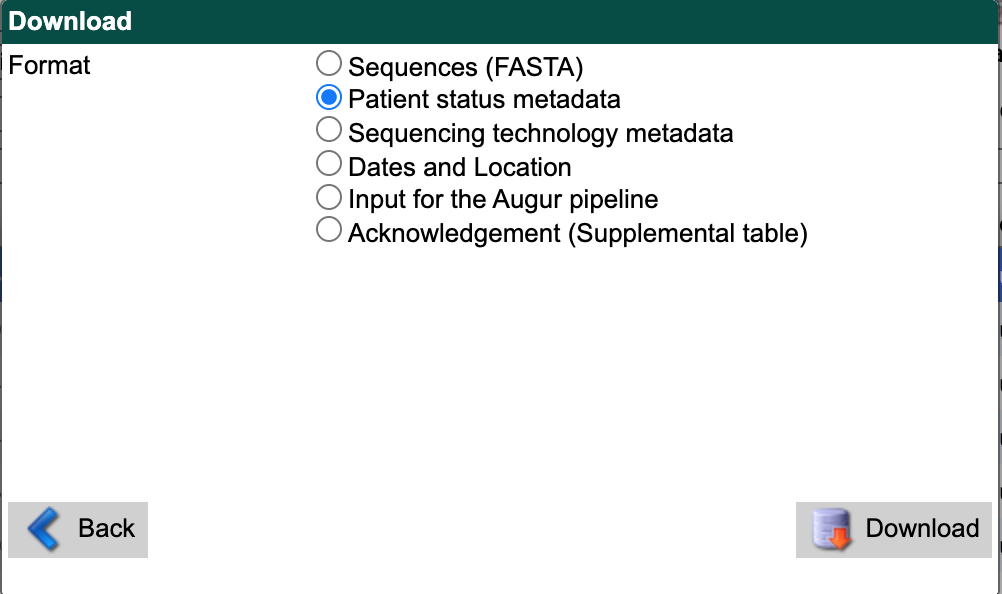
GISAID search download window showing “Patient status metadata” option
You can use these files as inputs for the workflow like so.
# Define inputs for the workflow.
inputs:
- name: washington
metadata: data/gisaid_washington_metadata.tsv
sequences: data/gisaid_washington_sequences.fasta
Download contextual data for your region of interest
Next, select the “Downloads” link from the EpiCoV navigation bar.
GISAID EpiCoV navigation bar with “Downloads” link
Scroll to the “Genomic epidemiology” section and select the “nextregions” button.
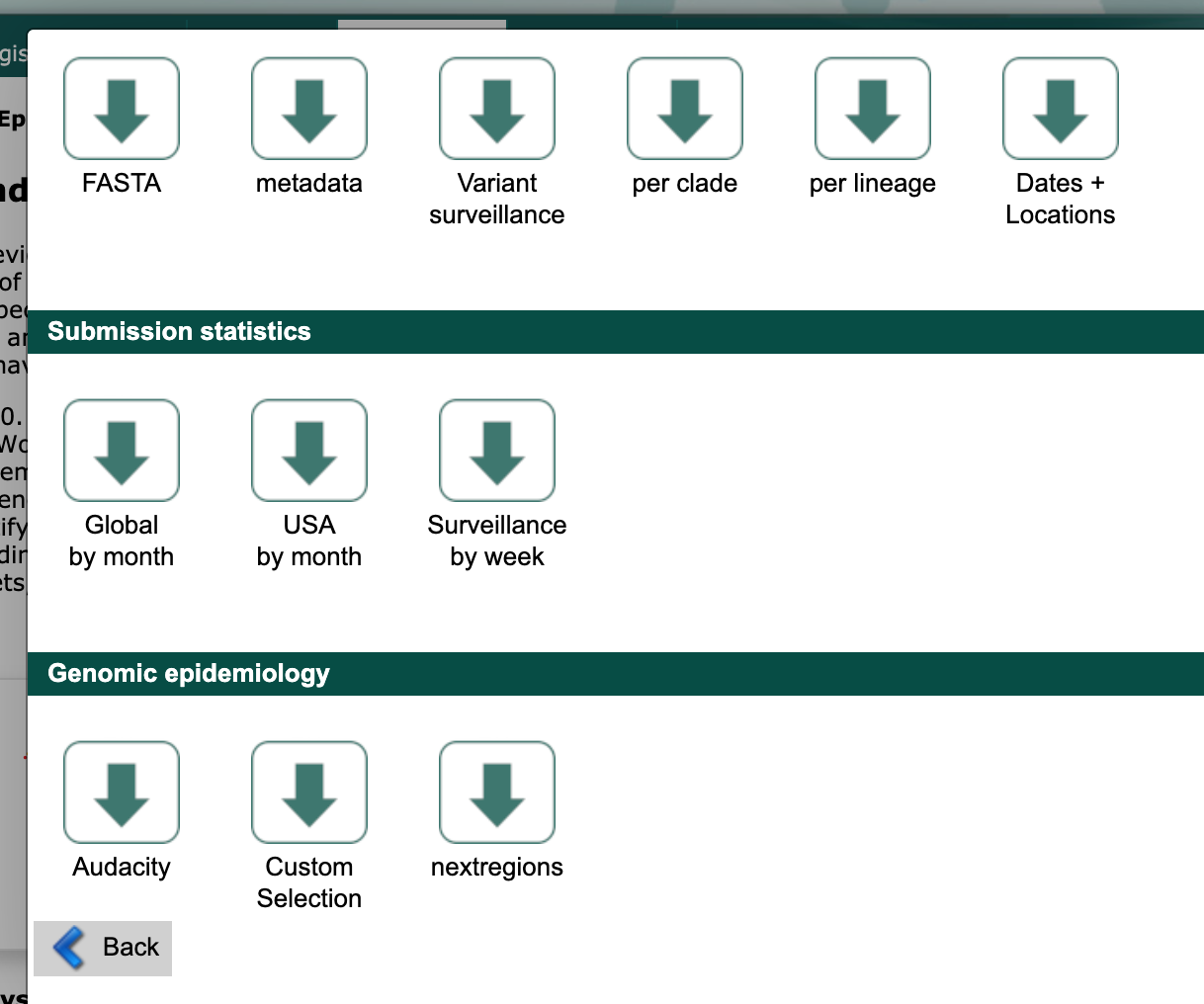
GISAID downloads window
Select the major region that corresponds to your region-specific data above (e.g., “North America”).
GISAID “nextregions” download window
Agree to the terms and conditions and download the corresponding file (named like ncov_north-america.tar.gz) to the data/ directory.
GISAID “nextregions” download terms and conditions
This compressed tar archive contains metadata and sequences corresponding to a recent Nextstrain build for that region with names like ncov_north-america.tsv and ncov_north-america.fasta, respectively. For example, the “North America” download contains data from Nextstrain’s North America build. These regional Nextstrain builds contain data from a specific region and contextual data from all other regions in the world. By default, GISAID provides these “nextregions” data in the “Input for the Augur pipeline” format.
As with the tar archive from the search results above, you can use the “nextregions” compressed tar archives as input to the Nextstrain workflow and the workflow will extract the appropriate contents for you. For example, you could update your inputs in the workflow config file from above to include the North American data as follows.
# Define inputs for the workflow.
inputs:
- name: washington
# The workflow will detect and extract the metadata and sequences
# from GISAID tar archives.
metadata: data/gisaid_washington.tar
sequences: data/gisaid_washington.tar
- name: north-america
# The workflow will similarly detect and extract metadata and
# sequences from compressed tar archives.
metadata: data/ncov_north-america.tar.gz
sequences: data/ncov_north-america.tar.gz
Alternately, you can extract the data from the compressed tar archive into the data/ directory.
tar zxvf data/ncov_north-america.tar.gz
You can use these extracted files as inputs for the workflow.
# Define inputs for the workflow.
inputs:
- name: washington
# The workflow will detect and extract the metadata and sequences
# from GISAID tar archives.
metadata: data/gisaid_washington.tar
sequences: data/gisaid_washington.tar
- name: north-america
# The workflow supports uncompressed or compressed input files.
metadata: data/ncov_north-america.tsv
sequences: data/ncov_north-america.fasta